R-loops: dynamic and widespread nucleic acid structures in the human genome
R-loops are three-stranded nucleic acid structures generated upon the hybridization of an RNA strand to a complementary DNA strand (Figure). This RNA:DNA hybrid forces the second DNA strand into a looped out state, hence the name of these structures.

Most available evidence suggest that R-loops are formed co-transcriptionally in cis although situations where the RNA strand invades a duplex DNA in trans have been reported.
One of the key factors predisposing to R-loop formation is positive GC skew – a measure of the distribution of guanine (G) and cytosine (C) residues across the two DNA strands. Positive GC skew denotes an asymmetry in this distribution such that more guanines are located on the “top” (5’-3’) strand than the other (see red / blue on the figure above). Positive GC skew enables the generation of a G-rich RNA upon transcription of a C-rich template DNA strand. Analyses of the stability of RNA:DNA duplexes reveal that G-rich RNAs base-paired to C-rich DNA strands are far more stable that the corresponding DNA:DNA duplex. The G-rich RNA strand therefore has a thermodynamic advantage for hybridization over the complementary DNA strand. During transcription, it is thought that the newly synthesized RNA strand, upon leaving the RNA exit channel of the traveling RNA polymerase complex, can compete with the non-template DNA strand for re-annealing to the template DNA strand (see Figures). This occurs as the transcription bubble closes down behind the transcription machinery. G-rich RNAs are thought to be more likely to out compete and ultimately displace the DNA strand owing to their ability to form stable RNA:DNA hybrid structures. Interestingly transcription that generates a C-rich RNA does not predispose to R-loop formation. Indeed C-rich RNA strands base-paired to G-rich DNA strands are poorly stable. Therefore R-loop formation is highly directional.
High-throughput genomics technologies to measure R-loop formation in cells
R-loop formation can easily be detected upon in vitro transcription of cloned, GC skewed, DNA templates. However, detecting R-loop formation in the genomes of live cells has remained difficult for years. This is due in part to the fact that these structures are likely to be transient in nature and may form at low steady-state frequencies.
Nonetheless we and others have developed independent methods to measure R-loop formation in the genomes of any organism. These methods are as follow:
- Non-denaturing bisulfite footprinting (1): this method makes use of the exquisite sensitivity of sodium bisulfite for single-stranded DNA. When applied to genomic DNA in a non-denaturing manner, sodium bisulfite will trigger C to U deamination at (unmethylated) cytosines that are unpaired. R-loops will appear as long patches of C to T conversion after PCR amplification of specific loci. These bisulfite sensitivity over conversion tracks can be removed if the genomic DNA is treated with Ribonuclease H, an enzyme that specifically degrades RNA:DNA hybrids, prior to bisulfite footprinting. See Ginno et al, (2012)2 for examples of R-loop footprinting in the human genome.
- Affinity purification of RNA:DNA hybrids (2): we recently made use of a catalytically-inactive but binding competent version of the human Ribonuclease H1 enzyme to affinity purify RNA:DNA hybrids and R-loops from the human genome. For this, the modified RNASEH1 protein fused to a Maltose-Binding protein tag was purified recombinantly and used to catch the structures. The recovered material was then analyzed using high-throughput sequencing in a technique we termed DRIVE-seq (DNA:RNA In Vitro Enrichment coupled to sequencing).
- DNA:RNA ImmunoPrecipitation (DRIP) (2): this methods takes advantage of the monoclonal S9.6 antibody which recognizes RNA:DNA hybrids with high specificity and little to no sequence-specificity. Immunoprecipitated material can easily be coupled to high-throughput sequening (DRIP-seq) to generate genome-wide maps of RNA:DNA hybrid formation. As expected, pre-treatment of the genome with Ribonuclease H abolishes one’s ability to immunoprecipitate any material, thereby demonstrating the specificity of the method. Given its low background noise and robustness, this method is becoming the method of choice for analysis of RNA:DNA hybrids.
Click here to access our DRIP-seq datasets.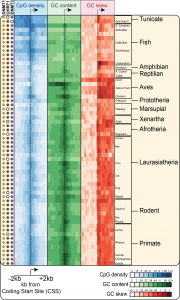
CpG islands, GC skew, and R-loops
The largest promoter class in Vertebrates corresponds to CpG island (CGI) promoters. CGI promoters are responsible for recruiting the transcription machinery and initiating transcription at ~60% of human genes, mostly genes with a “housekeeping” function that must be expressed at most times in most cell types. At the sequence level, CGIs have been historically defined as GC-rich regions that show a high density of CpG dinucleotides relative to the rest of the CpG-poor genome. Our work has shown that CGI promoters also show strong GC skew immediately downstream of the transcription start site. CGIs are therefore well-suited for R-loop formation and are indeed a hotspot of R-loop formation (2). Our recent work showed that GC skew at CGI promoters is conserved in most vertebrates, suggesting that R-loop formation at GC-skewed CGI promoters is a conserved feature of vertebrate genomes (3).
- Yu K, Chédin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003 May;4(5):442-51.[Link to Pubmed] [PDF]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012 Mar 30;45(6):814-25. doi: 10.1016/j.molcel.2012.01.017. Epub 2012 Mar 1.[Link to Pubmed] [PDF]
- Hartono S.R., Korf I.F., and Chédin* F. GC skew is a conserved property of unmethylated CpG island promoters across vertebrates. Nucleic Acids Research (in press). [Link to Pubmed]